An IFU Example - managing a discontiguous WCS¶
An IFU image represents the projection of several slices on a detector. Between the slices there are pixels which don’t belong to any slice. In general each slice has a unique WCS transform. There are two ways to represent this kind of transforms in GWCS depending on the way the instrument is calibrated and the available information.
Using a pixel to slice mapping¶
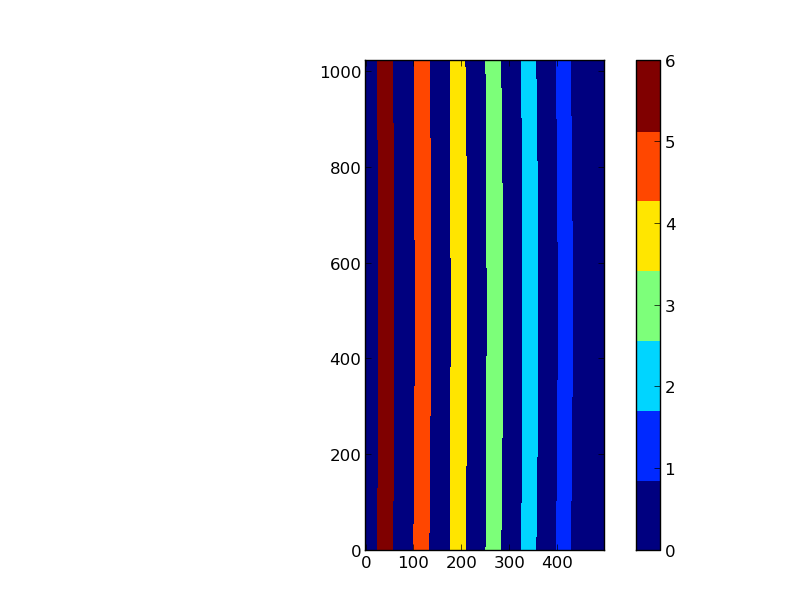
In this case a pixel map associating each pixel with a slice label (number or string) is available. The image below represents the projection of the slits of an IFU on a detector with a size (500, 1000). Slices are labeled from 1 to 6, while label 0 is reserved for pixels between the slices.
There are several models in GWCS which are useful in creating a WCS.
Given (x, y) pixel indices, LabelMapperArray returns labels (int or str)
associated with these indices. RegionsSelector
maps labels with transforms. It uses the LabelMapperArray to map
these transforms to pixel indices.
A step by step example of constructing the WCS for an IFU with 6 slits follows.
First, import the usual packages.
>>> import numpy as np
>>> from astropy.modeling import models
>>> from astropy import coordinates as coord
>>> from astropy import units as u
>>> from gwcs import wcs, selector
>>> from gwcs import coordinate_frames as cf
The output frame is common for all slits and is a composite frame with two subframes,
CelestialFrame and SpectralFrame.
>>> sky_frame = cf.CelestialFrame(name='icrs', reference_frame=coord.ICRS(), axes_order=(0, 2))
>>> spec_frame = cf.SpectralFrame(name='wave', unit=(u.micron,), axes_order=(1,), axes_names=('lambda',))
>>> cframe = cf.CompositeFrame([sky_frame, spec_frame], name='world')
>>> det = cf.Frame2D(name='detector')
All slices have the same input and output frames, however each slices has a different model transforming from pixels to world coordinates (RA, lambda, dec). For the sake of brevity this example uses a simple shift transform for each slice. Detailed examples of how to create more realistic transforms are available in Adding distortion to the imaging example.
>>> transforms = {}
>>> for i in range(1, 7):
... transforms[i] = models.Mapping([0, 0, 1]) | models.Shift(i * 0.1) & models.Shift(i * 0.2) & models.Scale(i * 0.1)
One way to initialize LabelMapperArray is to pass it the shape of the array and the vertices
of each slit on the detector {label: vertices} see :meth: from_vertices.
In this example the mask is an array with the size of the detector where each item in the array
corresponds to a pixel on the detector and its value is the slice number (label) this pixel
belongs to.
Assuming the array is stored in ASDF format, create the mask:
Create the pixel to world transform for the entire IFU:
>>> regions_transform = selector.RegionsSelector(inputs=['x','y'],
... outputs=['ra', 'dec', 'lam'],
... selector=transforms,
... label_mapper=mask,
... undefined_transform_value=np.nan)
The WCS object now can evaluate simultaneously the transforms of all slices.
>>> wifu = wcs.WCS(forward_transform=regions_transform, output_frame=cframe, input_frame=det)
>>> y, x = mask.mapper.shape
>>> y, x = np.mgrid[:y, :x]
>>> r, d, l = wifu(x, y)
or of single slices.
The set_input() method returns the forward_transform for
a specific label.
>>> wifu.forward_transform.set_input(4)(1, 2)
(1.4, 1.8, 0.8)
Custom model storing transforms in a dictionary¶
In case a pixel to slice mapping is not available, one can write a custom mdoel storing transforms in a dictionary. The model would look like this:
from astropy.modeling.core import Model
from astropy.modeling.parameters import Parameter
class CustomModel(Model):
inputs = ('label', 'x', 'y')
outputs = ('xout', 'yout')
def __init__(self, labels, transforms):
super().__init__()
self.labels = labels
self.models = models
def evaluate(self, label, x, y):
index = self.labels.index(label)
return self.models[index](x, y)